Project: Peptide aggregation on a solid surface
Relevant papers:
- Morriss-Andrews, A and Shea, J-E. "Kinetic pathways to peptide aggregation on surfaces: The effects of β-sheet propensity and surface attraction" J. Chem. Phys. 136, 065103 (2012)
- Morriss-Andrews, A, Bellesia, G, and Shea, J-E. "Effects of surface interactions on peptide aggregate morphology" J. Chem. Phys. 135, 085102 (2011)
We simulated peptide aggregation on a solid surface using the Shea coarse-grained peptide model. Recently, experimental setups include a variety of model surfaces such as mica, graphite, or Teflon. In some cases, ordered fibrils form on the surface where only amorphous oligomers appear in bulk solution.
We analyzed the kinetics and thermodynamics of how a solid surface modulates propensity toward the formation of fibrillar aggregates. At low surface attractions, we found multiple pathways leading to aggregate formation on the surface, depending on both the surface attraction experienced by the peptides and the peptides' chiral stiffness, which governs beta-sheet propensity in the bulk. A sufficiently strong surface attraction overrides the effect of chiral stiffness, leading to a single pathway (direct peptide deposition onto the surface) and a single aggregate morphology (single-layered fibrils).
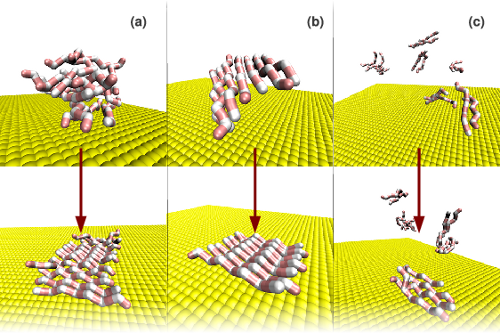
(a) amorphous-prone peptides and weaker surface attraction
(b) fibril-prone peptides and weaker surface attraction
(c) strong surface attraction (individual peptide deposition)
Thermodynamic simulations were conducted using the parallel tempering algorithm. We ran a range of temperatures of the same system, and exchanged the temperatures of the replicas according to a metropolis condition. This greatly enhances the sampling efficiency by incorporating a strategy from Monte Carlo simulation into molecular dynamics. It comes at the cost of losing kinetic information, since trajectories at a constant temperature are discontinuous. For this reason, we also conducted shorter, continuous simulations to elucidate the pathways of initial surface binding and aggregate formation.