Research
This work is done as part of my Ph.D., and is advised by Joan-Emma Shea at the University of California, Santa Barbara.
Projects
 Seeded Aggregation |
 Solid Surface |
 Lipid Bilayer |
 Polymer Chain |
General Info
Protein aggregation is a process whereby many proteins or peptides self-assemble into a single supramolecular structure. It is a topic of considerable biological interest due to its occurrence in several diseases, such as Alzheimer's and Parkinson's. In such pathological cases, mis-folded proteins become insoluble and aggregate into fibrillar structures known as amyloids.
Despite the considerable efforts of the scientific community, the physical mechanisms leading to protein aggregation are poorly understood. Experimentally, this is due to the extreme difficulty of resolving the process both spatially and temporally, exacerbated by the size and complexity of aggregates. Computer simulation sidesteps the issue of resolution, but runs into the barrier of computational complexity. Atomic detail simulations are in practice limited to the formation of small oligomers comprising only a few peptides, due to constraints in both system size and the aggregation time scale.
Coarse-graining is an effort to reduce the computational complexity of large systems. We use a united atom model where a group of mechanically-coupled atoms are represented by a single spherical particle. This reduces the effective number of atoms in the system, and allows for the simulation of larger systems on longer time scales.
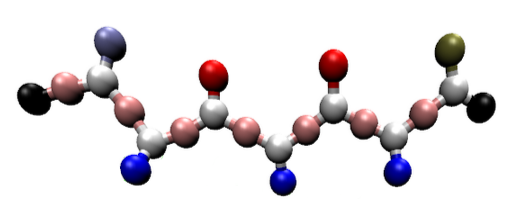
The Shea coarse-grained peptide model
We model short seven-residue peptides with a coarse-grained phenomenological model that exhibits a range of aggregate morphologies upon tuning a single parameter, the backbone chiral stiffness. At low values of this parameter, the peptides form amorphous, entropy-dominated oligomers; at high values, the peptides form multi-layered, energy-dominated fibrils. Intermediate values give rise to beta-barrels and mixed aggregate morphologies. Because our peptide model exhibits such rich structural diversity upon tuning a single parameter, it makes an excellent model system for studying the physical processes behind peptide aggregation.
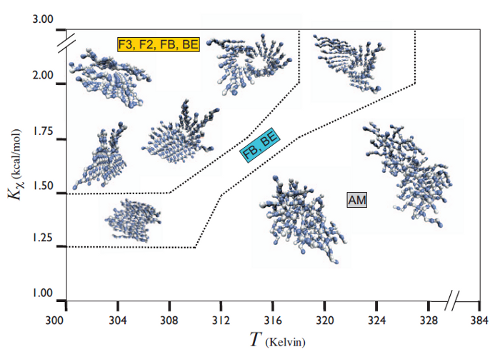
Bellesia and Shea, JCP 130, 145103 (2009)
Proteins can aggregate in bulk solution, however, aggregation in vivo occurs in the crowded environment of the cellular milieu, in particular, in the presence of surfaces such as membranes or large biomolecular assemblies. Our work focuses on peptide aggregation on three surfaces: the surface of a pre-existing fibril seed, a solid surface, and a lipid bilayer. Additionally, we are comparing the collapse of a polymer chain in explicit and implicit water.