Project: Peptide aggregation on a lipid bilayer
Relevant paper: Morriss-Andrews, A, Brown, FLH, and Shea, J-E. "A Coarse-Grained Model for Peptide Aggregation on a Membrane Surface" J. Phys. Chem. B. 118, (28), 8240 (2014)
This work was done in collaboration with the Brown group.
Many aggregates adsorb onto membranes. For instance, the amyloid-beta peptide, implicated in Alzheimer's disease, has a relatively high affinity to bind irreversibly to phospholipid membranes. There are several models by which protein aggregates are theorized to cause membrane damage: (a) fibrils carpet the membrane leading to a pressure differential between leaflets, (b) amyloids embed into the bilayer causing pore formation, and (c) detergent effects thin the membrane as lipids bind preferentially to the aggregate instead of each other.
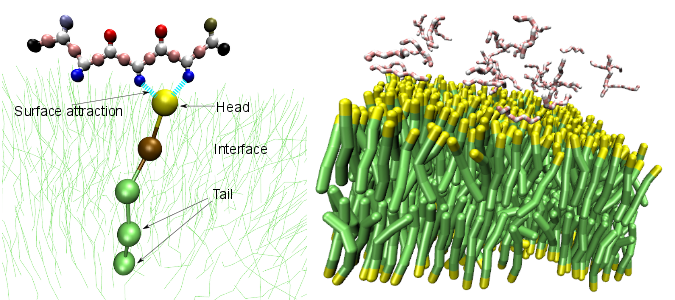
Left: One lipid of the bilayer is singled out; Right: An initial condition of full system.
We are simulating the aggregation of the Shea coarse-grained peptide model onto the surface of a lipid membrane. Our results highlight the importance of membrane fluidity in the kinetics of aggregate formation and the aggregates' final morphologies, contrasting our earlier study of peptide aggregation on a solid surface.
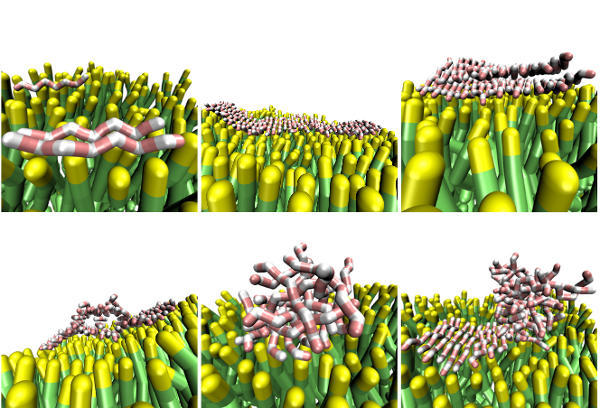
Bilayer fluctuations cause aggregates to bind less tightly to the membrane than to a solid surface, allowing for less flat morphologies, such as beta-barrels. Conversely, we observe fibrillar peptide aggregates locally dampening membrane fluctuations, and causing lipids underneath fibrils temporarily to adopt a pseudo-lattice structure complementary to the fibril.